!pip install -q torch transformers transformers accelerate bitsandbytes langchain sentence-transformers faiss-gpu openpyxl pacmapAdvanced RAG
This notebook demonstrates how you can build an advanced RAG (Retrieval Augmented Generation) for answering a user’s question about a specific knowledge base (here, the HuggingFace documentation), using LangChain.
For an introduction to RAG, you can check this other cookbook!
RAG systems are complex, with many moving parts: here a RAG diagram, where we noted in blue all possibilities for system enhancement:
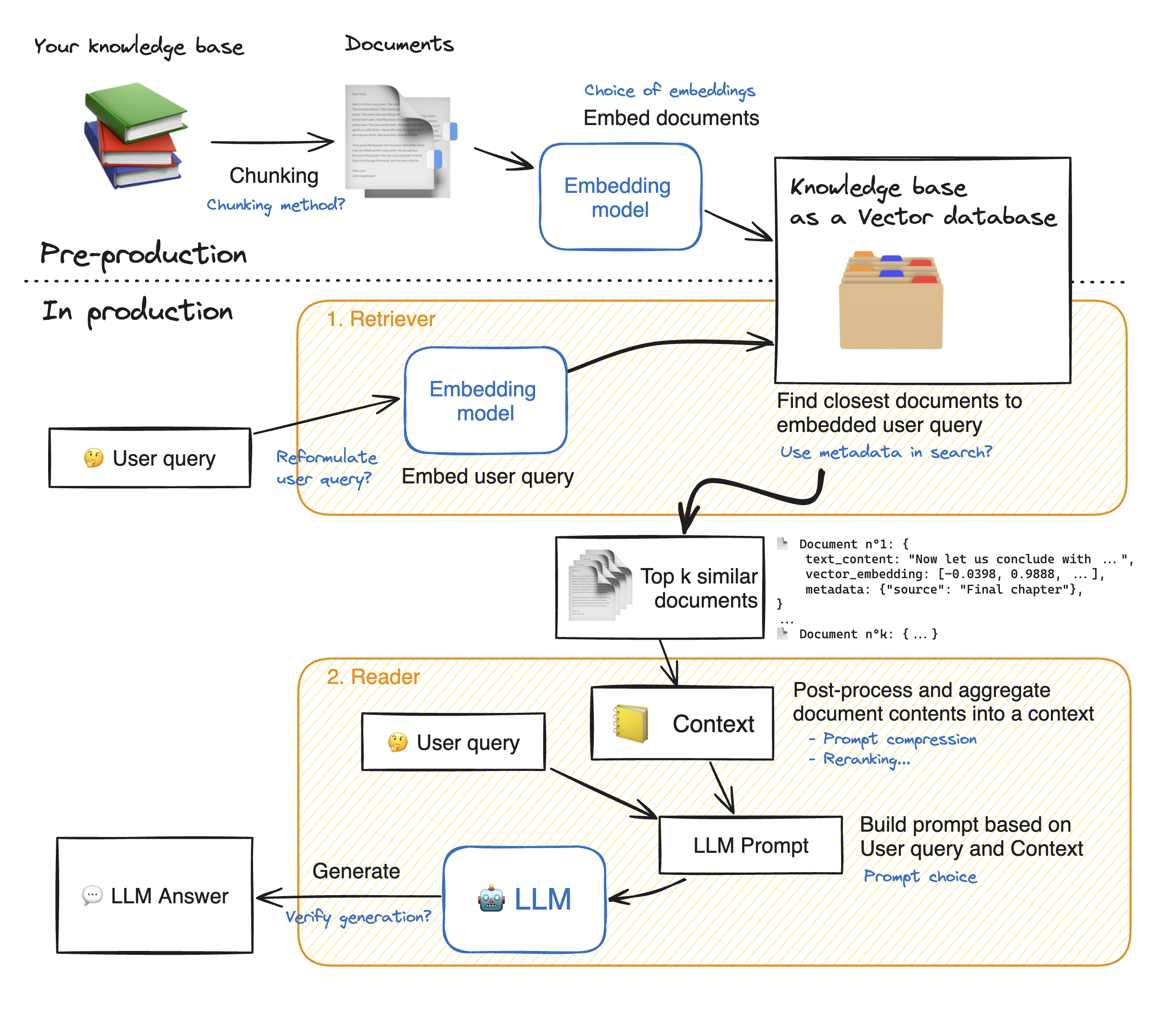
💡 As you can see, there are many steps to tune in this architecture: tuning the system properly will yield significant performance gains.
In this notebook, we will take a look into many of these blue notes to see how to tune your RAG system and get the best performance.
Let’s dig into the model building! First, we install the required model dependancies.
%reload_ext dotenv
%dotenvfrom tqdm.notebook import tqdm
import pandas as pd
from typing import Optional, List, Tuple
from datasets import Dataset
import matplotlib.pyplot as plt
pd.set_option(
"display.max_colwidth", None
) - 1
- This will be helpful when visualizing retriever outputs
Load your knowledge base
import datasets
ds = datasets.load_dataset("m-ric/huggingface_doc", split="train")from langchain.docstore.document import Document as LangchainDocument
RAW_KNOWLEDGE_BASE = [
LangchainDocument(page_content=doc["text"], metadata={"source": doc["source"]})
for doc in tqdm(ds)
]1. Retriever - embeddings 🗂️
The retriever acts like an internal search engine: given the user query, it returns a few relevant snippets from your knowledge base.
These snippets will then be fed to the Reader Model to help it generate its answer.
So our objective here is, given a user question, to find the most snippets from our knowledge base to answer that question.
This is a wide objective, it leaves open some questions. How many snippets should we retrieve? This parameter will be named top_k.
How long should these snippets be? This is called the chunk size. There’s no one-size-fits-all answers, but here are a few elements: - 🔀 Your chunk size is allowed to vary from one snippet to the other. - Since there will always be some noise in your retrieval, increasing the top_k increases the chance to get relevant elements in your retrieved snippets. 🎯 Shooting more arrows increases your probability to hit your target. - Meanwhile, the summed length of your retrieved documents should not be too high: for instance, for most current models 16k tokens will probably drown your Reader model in information due to Lost-in-the-middle phenomenon. 🎯 Give your reader model only the most relevant insights, not a huge pile of books!
In this notebook, we use Langchain library since it offers a huge variety of options for vector databases and allows us to keep document metadata throughout the processing.
1.1 Split the documents into chunks
- In this part, we split the documents from our knowledge base into smaller chunks which will be the snippets on which the reader LLM will base its answer.
- The goal is to prepare a collection of semantically relevant snippets. So their size should be adapted to precise ideas: too small will truncate ideas, too large will dilute them.
💡 Many options exist for text splitting: splitting on words, on sentence boundaries, recursive chunking that processes documents in a tree-like way to preserve structure information… To learn more about chunking, I recommend you read this great notebook by Greg Kamradt.
- Recursive chunking breaks down the text into smaller parts step by step using a given list of separators sorted from the most important to the least important separator. If the first split doesn’t give the right size or shape chunks, the method repeats itself on the new chunks using a different separator. For instance with the list of separators
["\n\n", "\n", ".", ""]:- The method will first break down the document wherever there is a double line break
"\n\n". - Resulting documents will be split again on simple line breaks
"\n", then on sentence ends".". - And finally, if some chunks are still too big, they will be split whenever they overflow the maximum size.
- The method will first break down the document wherever there is a double line break
- With this method, the global structure is well preserved, at the expense of getting slight variations in chunk size.
This space lets you visualize how different splitting options affect the chunks you get.
🔬 Let’s experiment a bit with chunk sizes, beginning with an arbitrary size, and see how splits work. We use Langchain’s implementation of recursive chunking with RecursiveCharacterTextSplitter. - Parameter chunk_size controls the length of individual chunks: this length is counted by default as the number of characters in the chunk. - Parameter chunk_overlap lets adjacent chunks get a bit of overlap on each other. This reduces the probability that an idea could be cut in half by the split between two adjacent chunks. We ~arbitrarily set this to 1/10th of the chunk size, you could try different values!
from langchain.text_splitter import RecursiveCharacterTextSplitter
# We use a hierarchical list of separators specifically tailored for splitting Markdown documents
# This list is taken from LangChain's MarkdownTextSplitter class.
MARKDOWN_SEPARATORS = [
"\n#{1,6} ",
"```\n",
"\n\\*\\*\\*+\n",
"\n---+\n",
"\n___+\n",
"\n\n",
"\n",
" ",
"",
]
text_splitter = RecursiveCharacterTextSplitter(
chunk_size=1000,
chunk_overlap=100,
add_start_index=True,
strip_whitespace=True,
separators=MARKDOWN_SEPARATORS,
)
docs_processed = []
for doc in RAW_KNOWLEDGE_BASE:
docs_processed += text_splitter.split_documents([doc])- 1
- The maximum number of characters in a chunk: we selected this value arbitrally
- 2
- The number of characters to overlap between chunks
- 3
-
If
True, includes chunk’s start index in metadata - 4
-
If
True, strips whitespace from the start and end of every document
We also have to keep in mind that when embedding documents, we will use an embedding model that has accepts a certain maximum sequence length max_seq_length.
So we should make sure that our chunk sizes are below this limit, because any longer chunk will be truncated before processing, thus losing relevancy.
from sentence_transformers import SentenceTransformer
# To get the value of the max sequence_length, we will query the underlying `SentenceTransformer` object used in the RecursiveCharacterTextSplitter.
print(
f"Model's maximum sequence length: {SentenceTransformer('thenlper/gte-small').max_seq_length}"
)
from transformers import AutoTokenizer
tokenizer = AutoTokenizer.from_pretrained("thenlper/gte-small")
lengths = [len(tokenizer.encode(doc.page_content)) for doc in tqdm(docs_processed)]
# Plot the distrubution of document lengths, counted as the number of tokens
fig = pd.Series(lengths).hist()
plt.title("Distribution of document lengths in the knowledge base (in count of tokens)")
plt.show()👀 As you can see, the chunk lengths are not aligned with our limit of 512 tokens, and some documents are above the limit, thus some part of them will be lost in truncation! - So we should change the RecursiveCharacterTextSplitter class to count length in number of tokens instead of number of characters. - Then we can choose a specific chunk size, here we would choose a lower threshold than 512: - smaller documents could allow the split to focus more on specific ideas. - But too small chunks would split sentences in half, thus losing meaning again: the proper tuning is a matter of balance.
from langchain.text_splitter import RecursiveCharacterTextSplitter
from transformers import AutoTokenizer
EMBEDDING_MODEL_NAME = "thenlper/gte-small"
def split_documents(
chunk_size: int,
knowledge_base: List[LangchainDocument],
tokenizer_name: Optional[str] = EMBEDDING_MODEL_NAME,
) -> List[LangchainDocument]:
"""
Split documents into chunks of maximum size `chunk_size` tokens and return a list of documents.
"""
text_splitter = RecursiveCharacterTextSplitter.from_huggingface_tokenizer(
AutoTokenizer.from_pretrained(tokenizer_name),
chunk_size=chunk_size,
chunk_overlap=int(chunk_size / 10),
add_start_index=True,
strip_whitespace=True,
separators=MARKDOWN_SEPARATORS,
)
docs_processed = []
for doc in knowledge_base:
docs_processed += text_splitter.split_documents([doc])
# Remove duplicates
unique_texts = {}
docs_processed_unique = []
for doc in docs_processed:
if doc.page_content not in unique_texts:
unique_texts[doc.page_content] = True
docs_processed_unique.append(doc)
return docs_processed_unique
docs_processed = split_documents(
512, # We choose a chunk size adapted to our model
RAW_KNOWLEDGE_BASE,
tokenizer_name=EMBEDDING_MODEL_NAME,
)
# Let's visualize the chunk sizes we would have in tokens from a common model
from transformers import AutoTokenizer
tokenizer = AutoTokenizer.from_pretrained(EMBEDDING_MODEL_NAME)
lengths = [len(tokenizer.encode(doc.page_content)) for doc in tqdm(docs_processed)]
fig = pd.Series(lengths).hist()
plt.title("Distribution of document lengths in the knowledge base (in count of tokens)")
plt.show()➡️ Now the chunk length distribution looks better!
1.2 Building the vector database
We want to compute the embeddings for all the chunks of our knowledge base: to learn more on sentence embeddings, we recommend reading this guide.
How does retrieval work ?
Once the chunks are all embedded, we store them into a vector database. When the user types in a query, it gets embedded by the same model previously used, and a similarity search returns the closest documents from the vector database.
The technical challenge is thus, given a query vector, to quickly find the nearest neighbours of this vector in the vector database. To do this, we need to choose two things: a distance, and a search algorithm to find the nearest neighbors quickly within a database of thousands of records.
Nearest Neighbor search algorithm
There are plentiful choices for the nearest neighbor search algorithm: we go with Facebook’s FAISS, since FAISS is performant enough for most use cases, and it is well known thus widely implemented.
Distances
Regarding distances, you can find a good guide here. In short:
- Cosine similarity computes similarity between two vectors as the cosinus of their relative angle: it allows us to compare vector directions are regardless of their magnitude. Using it requires to normalize all vectors, to rescale them into unit norm.
- Dot product takes into account magnitude, with the sometimes undesirable effect that increasing a vector’s length will make it more similar to all others.
- Euclidean distance is the distance between the ends of vectors.
You can try this small exercise to check your understanding of these concepts. But once vectors are normalized, the choice of a specific distance does not matter much.
Our particular model works well with cosine similarity, so choose this distance, and we set it up both in the Embedding model, and in the distance_strategy argument of our FAISS index. With cosine similarity, we have to normalize our embeddings.
🚨👇 The cell below takes a few minutes to run on A10G!
from langchain.vectorstores import FAISS
from langchain_community.embeddings import HuggingFaceEmbeddings
from langchain_community.vectorstores.utils import DistanceStrategy
embedding_model = HuggingFaceEmbeddings(
model_name=EMBEDDING_MODEL_NAME,
multi_process=True,
model_kwargs={"device": "cuda"},
encode_kwargs={"normalize_embeddings": True}, # set True for cosine similarity
)
KNOWLEDGE_VECTOR_DATABASE = FAISS.from_documents(
docs_processed, embedding_model, distance_strategy=DistanceStrategy.COSINE
)👀 To visualize the search for the closest documents, let’s project our embeddings from 384 dimensions down to 2 dimensions using PaCMAP.
💡 We chose PaCMAP rather than other techniques such as t-SNE or UMAP, since it is efficient (preserves local and global structure), robust to initialization parameters and fast.
# embed a user query in the same space
user_query = "How to create a pipeline object?"
query_vector = embedding_model.embed_query(user_query)import pacmap
import numpy as np
import plotly.express as px
embedding_projector = pacmap.PaCMAP(
n_components=2, n_neighbors=None, MN_ratio=0.5, FP_ratio=2.0, random_state=1
)
embeddings_2d = [
list(KNOWLEDGE_VECTOR_DATABASE.index.reconstruct_n(idx, 1)[0])
for idx in range(len(docs_processed))
] + [query_vector]
# fit the data (The index of transformed data corresponds to the index of the original data)
documents_projected = embedding_projector.fit_transform(np.array(embeddings_2d), init="pca")df = pd.DataFrame.from_dict(
[
{
"x": documents_projected[i, 0],
"y": documents_projected[i, 1],
"source": docs_processed[i].metadata["source"].split("/")[1],
"extract": docs_processed[i].page_content[:100] + "...",
"symbol": "circle",
"size_col": 4,
}
for i in range(len(docs_processed))
]
+ [
{
"x": documents_projected[-1, 0],
"y": documents_projected[-1, 1],
"source": "User query",
"extract": user_query,
"size_col": 100,
"symbol": "star",
}
]
)
# visualize the embedding
fig = px.scatter(
df,
x="x",
y="y",
color="source",
hover_data="extract",
size="size_col",
symbol="symbol",
color_discrete_map={"User query": "black"},
width=1000,
height=700,
)
fig.update_traces(
marker=dict(opacity=1, line=dict(width=0, color="DarkSlateGrey")), selector=dict(mode="markers")
)
fig.update_layout(
legend_title_text="<b>Chunk source</b>",
title="<b>2D Projection of Chunk Embeddings via PaCMAP</b>",
)
fig.show()
➡️ On the graph above, you can see a spatial representation of the kowledge base documents. As the vector embeddings represent the document’s meaning, their closeness in meaning should be reflected in their embedding’s closeness.
The user query’s embedding is also shown : we want to find the k document that have the closest meaning, thus we pick the k closest vectors.
In the LangChain vector database implementation, this search operation is performed by the method vector_database.similarity_search(query).
Here is the result:
print(f"\nStarting retrieval for {user_query=}...")
retrieved_docs = KNOWLEDGE_VECTOR_DATABASE.similarity_search(query=user_query, k=5)
print("\n==================================Top document==================================")
print(retrieved_docs[0].page_content)
print("==================================Metadata==================================")
print(retrieved_docs[0].metadata)2. Reader - LLM 💬
In this part, the LLM Reader reads the retrieved context to formulate its answer.
There are actually substeps that can all be tuned: 1. The content of the retrieved documents is aggregated together into the “context”, with many processing options like prompt compression. 2. The context and the user query are aggregated into a prompt then given to the LLM to generate its answer.
2.1. Reader model
The choice of a reader model is important on a few aspects: - the reader model’s max_seq_length must accomodate our prompt, which includes the context output by the retriever call: the context consists in 5 documents of 512 tokens each, so we aim for a context length of 4k tokens at least. - the reader model
For this example, we chose HuggingFaceH4/zephyr-7b-beta, a small but powerful model.
With many models being released every week, you may want to substitute this model to the latest and greatest. The best way to keep track of open source LLMs is to check the Open-source LLM leaderboard.
To make inference faster, we will load the quantized version of the model:
from transformers import pipeline
import torch
from transformers import AutoTokenizer, AutoModelForCausalLM, BitsAndBytesConfig
READER_MODEL_NAME = "HuggingFaceH4/zephyr-7b-beta"
bnb_config = BitsAndBytesConfig(
load_in_4bit=True,
bnb_4bit_use_double_quant=True,
bnb_4bit_quant_type="nf4",
bnb_4bit_compute_dtype=torch.bfloat16,
)
model = AutoModelForCausalLM.from_pretrained(READER_MODEL_NAME, quantization_config=bnb_config)
tokenizer = AutoTokenizer.from_pretrained(READER_MODEL_NAME)
READER_LLM = pipeline(
model=model,
tokenizer=tokenizer,
task="text-generation",
do_sample=True,
temperature=0.2,
repetition_penalty=1.1,
return_full_text=False,
max_new_tokens=500,
)READER_LLM("What is 4+4? Answer:")2.2. Prompt
The RAG prompt template below is what we will feed to the Reader LLM: it is important to have it formatted in the Reader LLM’s chat template.
We give it our context and the user’s question.
prompt_in_chat_format = [
{
"role": "system",
"content": """Using the information contained in the context,
give a comprehensive answer to the question.
Respond only to the question asked, response should be concise and relevant to the question.
Provide the number of the source document when relevant.
If the answer cannot be deduced from the context, do not give an answer.""",
},
{
"role": "user",
"content": """Context:
{context}
---
Now here is the question you need to answer.
Question: {question}""",
},
]
RAG_PROMPT_TEMPLATE = tokenizer.apply_chat_template(
prompt_in_chat_format, tokenize=False, add_generation_prompt=True
)
print(RAG_PROMPT_TEMPLATE)Let’s test our Reader on our previously retrieved documents!
retrieved_docs_text = [
doc.page_content for doc in retrieved_docs
] # we only need the text of the documents
context = "\nExtracted documents:\n"
context += "".join([f"Document {str(i)}:::\n" + doc for i, doc in enumerate(retrieved_docs_text)])
final_prompt = RAG_PROMPT_TEMPLATE.format(
question="How to create a pipeline object?", context=context
)
# Redact an answer
answer = READER_LLM(final_prompt)[0]["generated_text"]
print(answer)2.3. Reranking
A good option for RAG is to retrieve more documents than you want in the end, then rerank the results with a more powerful retrieval model before keeping only the top_k.
For this, Colbertv2 is a great choice: instead of a bi-encoder like our classical embedding models, it is a cross-encoder that computes more fine-grained interactions between the query tokens and each document’s tokens.
It is easily usable thanks to the RAGatouille library.
from ragatouille import RAGPretrainedModel
RERANKER = RAGPretrainedModel.from_pretrained("colbert-ir/colbertv2.0")3. Assembling it all!
from transformers import Pipeline
def answer_with_rag(
question: str,
llm: Pipeline,
knowledge_index: FAISS,
reranker: Optional[RAGPretrainedModel] = None,
num_retrieved_docs: int = 30,
num_docs_final: int = 5,
) -> Tuple[str, List[LangchainDocument]]:
# Gather documents with retriever
print("=> Retrieving documents...")
relevant_docs = knowledge_index.similarity_search(query=question, k=num_retrieved_docs)
relevant_docs = [doc.page_content for doc in relevant_docs] # keep only the text
# Optionally rerank results
if reranker:
print("=> Reranking documents...")
relevant_docs = reranker.rerank(question, relevant_docs, k=num_docs_final)
relevant_docs = [doc["content"] for doc in relevant_docs]
relevant_docs = relevant_docs[:num_docs_final]
# Build the final prompt
context = "\nExtracted documents:\n"
context += "".join([f"Document {str(i)}:::\n" + doc for i, doc in enumerate(relevant_docs)])
final_prompt = RAG_PROMPT_TEMPLATE.format(question=question, context=context)
# Redact an answer
print("=> Generating answer...")
answer = llm(final_prompt)[0]["generated_text"]
return answer, relevant_docsLet’s see how our RAG pipeline answers a user query.
question = "how to create a pipeline object?"
answer, relevant_docs = answer_with_rag(
question, READER_LLM, KNOWLEDGE_VECTOR_DATABASE, reranker=RERANKER
)print("==================================Answer==================================")
print(f"{answer}")
print("==================================Source docs==================================")
for i, doc in enumerate(relevant_docs):
print(f"Document {i}------------------------------------------------------------")
print(doc)✅ We now have a fully functional, performant RAG sytem. That’s it for today! Congratulations for making it to the end 🥳
To go further 🗺️
This is not the end of the journey! You can try many steps to improve your RAG system. We recommend doing so in an iterative way: bring small changes to the system and see what improves performance.
Setting up an evaluation pipeline
- 💬 “You cannot improve the model performance that you do not measure”, said Gandhi… or at least Llama2 told me he said it. Anyway, you should absolutely start by measuring performance: this means building a small evaluation dataset, then monitor the performance of your RAG system on this evaluation dataset.
Improving the retriever
🛠️ You can use these options to tune the results:
- Tune the chunking method:
- Size of the chunks
- Method: split on different separators, use semantic chunking…
- Change the embedding model
👷♀️ More could be considered: - Try another chunking method, like semantic chunking - Change the index used (here, FAISS) - Query expansion: reformulate the user query in slightly different ways to retrieve more documents.
Improving the reader
🛠️ Here you can try the following options to improve results: - Tune the prompt - Switch reranking on/off - Choose a more powerful reader model
💡 Many options could be considered here to further improve the results: - Compress the retrieved context to keep only the most relevant parts to answer the query. - Extend the RAG system to make it more user-friendly: - cite source - make conversational